About OHBM BrainHack 2020
At the OHBM Brainhack members of the community gather to work collaboratively on common projects.
The event also features a TrainTrack in which tutorials on open science practices are provided.
Brainhacks differ from typical academic conference in that attendees can actively take part in the program and co-learn from each other. Thus, they will be actively trying to create something rather than passively attend. Although many brainhack projects might involve coding, it is not a "requirement and many projects can be done without coding skills.
Share Ideas

Share your research and curiosity with other neuroimaging and computer science researchers.
Build Projects

Try the presented tools on provided data or bring your own tools/data!
Be social
Get to meet and socialize with people from all over the world!
Learn Tools

Learn about brain imaging data and how to apply the latest computational tools to your data.
What's Up? #OHBMHackathon
HackTrack projects

Learn and Enhance BrainIAK Tutorials: From Basics to Advanced fMRI Analyses
Hub : Americas
PROJECT DESCRIPTION
This project will help participants learn fMRI analyses and enhance BrainIAK Tutorials (Kumar et al., 2020). Participants can practice a tutorial covering any topic of interest, from basics to advanced techniques. They can also contribute to the tutorials, by making edits, fixing bugs, or enhancing them by applying the techniques to their own datasets. We have a list an issue list for the tutorials that participants can pick an issue and work on. Some of the advanced techniques included are: Inter-Subject Correlations (ISC) (Hasson et al., 2004; Simony et al., 2016), Shared Response Modeling (SRM) (Chen et al., 2015), and Event Segmentation methods (Baldassano et al., 2017)
More information in this github issue

BIDS to NiftyTorch: data preparation for deep learning
Hub : Americas
PROJECT DESCRIPTION
We aim to build an automated pipeline to make a copy of BIDS formatted project for the purpose of deep learning and automatically evaluate the dataset. This includes, creating a randomly selected sets of training, validation and test dataset for deep learning, searching for missing data, evaluating consistency across image input using image size and modality, and maybe adding a module for quality assessment and preprocessing of the input data.
More information in this github issue
Tracktools
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
In the latest years, I have developed/collected some tools for brain tractography, from inclusion in html, to 3Dprinter conversion to other manipulations. Codes is mostly in matlab and we should move in Python, moreover the converter from Trackvis to STL for 3Dprinting or Augmented Reality is not good. I am open minded to other ideas for tractography.
More information in this github issue
VB Index BIDS support / Pretty GUI
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
I have been collaborating / leading a project that produced some software to 1) create 'cortical gradients' and a simple edge detection technique called the VB Index (see preprint). We have a python and matlab tool but are focussing on the python version. The brainhack project will be simple, allow the toolbox to accept BIDS data and / or create a simple (but pretty) GUI.
More information in this github issue

Tools for Combatting Citation Bias
Hub : Americas
PROJECT DESCRIPTION
Recent work in neuroscience and other fields has identified a bias in citation practices such that papers from women and other minorities are under-cited relative to the number of such papers in the field. For this project, we seek to further the development of two tools to help researchers combat these trends in their own citation practices. The first tool is an interactive jupyter notebook that provides probabilistic gender break down of first and last authors from a bibliography in a bibtex file. The second is a Google Chrome extension that displays the probabilistic gender of first and last authors of papers on the common search engines Google Scholar and PubMed.
More information in this github issue
Wacky brain explosions Vol II.
Hub : Americas
PROJECT DESCRIPTION
Are you asking to yourself 'how can the year 2020 get more interesting?. The answer is simple: by using a particle simulator to explode some brains at the comfort of your home, in a completely safe virtual environment.
After concluding my previous hackaton project with 'more research is needed', here I am planning to continue exploding more brains in many of ways. All in the name of neuro-science, of course!
More information in this github issue

shimming toolbox: Ecosystem for real-time shimming with MRI
Hub : Americas
PROJECT DESCRIPTION
A prerequisite to correcting/shimming the MR 'B0' field is of course to measure it (commonly done using a dual-echo gradient-echo sequence). A recent paper by Fatnassi and Zaidi suggests that multi-echo (n>2) sequences may yield better accuracy and higher SNR. This brainhack project aims to implement multi-echo field mapping algorithms and assess their efficiency.
More information in this github issue
Hopfield Network Library
Hub : Americas
PROJECT DESCRIPTION
The goal of this project is to create an easy-to-use library for undirected neural networks, including Hopfield networks, Boltzmann machines, and Deep Belief networks. We also want to run empirical studies on the behavior of Hopfield networks, including memory capacity and time until convergence.
More information in this github issue
Vector Embeddings of Brain Dynamics
Hub : Americas
PROJECT DESCRIPTION
Whereas we can build time-varying functional connectivity (TVFC) graphs easily and efficiently using instantaneous wavelet coherence. And whereas we can compare TVFC graphs using a host of graph-graph metrics---from simple edge-wise overlap, to more robust methods from topological data analysis. The capacity to understand how TVFC states compare among volunteers is limited by the transductive nature of state-of-the-art manifold embedding algorithms such as tSNE and UMAP. Transductive methods tend to overweight the temporal adjacencies of graph-graph distances measured within single volunteers, and thereby ignore the large-scale graph-graph similarities measured across volunteers. To address this issue, this project aims to build vector embeddings of several TVFC graph-graph distances using a Siamese neural network.
More information in this github issue

Mapping the parameter space in EEG/MEG analyses
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
The parameter space of M/EEG analysis pipelines is like a jungle. We will map it! We will first identify and classify available pipelines (based on available pre-registrations) and identify common challenges researchers face, then come up with a semantic framework to create and compare different pipelines, map software options onto the different analysis steps, and finally start designing an adaptive app for M/EEG pre-registration!
More information in this github issue

Tutorial for cross decomposition algorithms (e.g. CCA, PLS) in Python
Hub : Asia / Pacific
PROJECT DESCRIPTION
Cross decomposition algorithms look for the relations between two (or more) blocks of variables. These methods are particularly used in neuroimaging to analyze associations between physiological/behavioral variables and brain structure/function. Between unsupervised and supervised modeling, this family of algorithms has many members (e.g. CCA, PLS regression, PLS canonical, PLS-PM, etc.) and many approaches are possible to validate the trained model (e.g. cross validation, bootstrapping, permutation test, etc.). In this project, I propose to write several Python tutorials to help the application and interpretation of these models in practice.
More information in this github issue

git-annex special remote for the open science framework
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
The Open Science Framework (OSF) is an amazing infrastructure for open science. In this project, we attempt to create a git-annex special remote implementation to leverage OSF filestorage and make the data storing that OSF provides even more useful. The git-annex OFS special remote would allow to transform OSF storage into git-annex repositories. Files in OSF storage could thus be consumed or exported fast and easily via git-annex or datalad, and published to repository-hosting services (GitHub, GitLab, Bitbucket, ...) as lightweight repositories that constitute an alternative access to the data stored on the OSF - that is: you can git clone a repository from for example GitHub and get the data from the OSF from the command line or in your scripts.
More information in this github issue

Multi-modality neurofeedback and biofeedback to treat children with ADHD or anxiety
Hub : Americas
PROJECT DESCRIPTION
Children with ADHD, PTSD or generalized anxiety benefit from neurofeedback from an EEG while performing a task (e.g. focusing, calming) to develop cognitive skills that can assist them to cope and perform better throughout life. Some problems with this training a) the tools need to be easy to access and setup (e.g. wearable); b) the feedback should be engaging and ideally involving multiple senses (e.g. visual, auditory, 3D, aromas); and c) the feedback should be based on both brain waves (EEG) and other indicators (e.g. body motion, heart rate, breathing rate) to provide richer self-understanding.
Problem: Develop an app for use with a wearable neurofeedback system (e.g. Muse, Emotiv, OpenBCI) that provides richer environmental feedback and/or sensing to better serve the ADHD, PTSD or generalized anxiety pediatric populations.
More information in this github issue

Bidsme -- bidsifier for multimodal datasets
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
One of the great ideas behind BIDS is unification of various modalities in the same dataset, sharing common metadata and providing a self-sufficient dataset ready to analyse. However various bidsificators target only one or two modalities. Bidsme project has been developed to provide a bidsificator as universal as possible, both in modalities and data formats. The project is based on two main ideas:
1. everything that is laboratory-depending (log files, events, demographic info, psychological assessments, format conversions) must be treated via plugins.
2. Data treated in object-oriented way: master class do the bidsifications, modality sub-class defines the BIDS-related parameters (entities, JSON fields etc...), data-format sub-sub-class implements how to read metadata.
Using this approach it is possible to integrate a new modality from scratch in one week, and fully create plugins for a given dataset in few days.
More information in this github issue
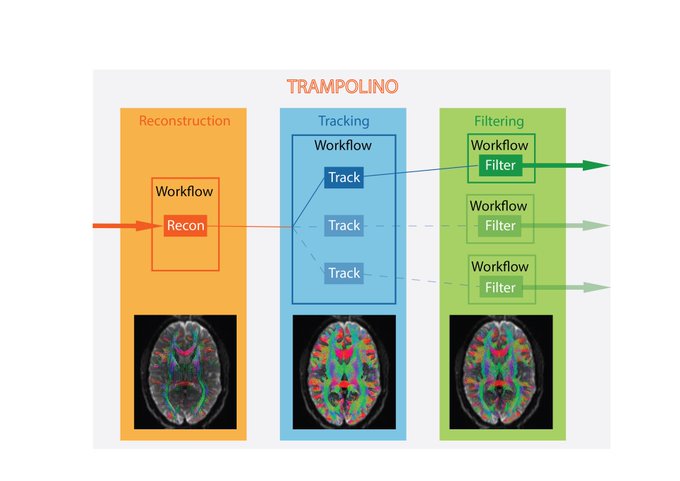
TRAMPOLINO: the Swiss Army Knife for tractography
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
I wrote TRAMPOLINO during BrainHack School with the idea of building a tool to easily do tractography across different packages. My vision now is to make it become the Swiss Army Knife for tractography: a tool able to try out things, even when you do not have the right software installed (!!!). The dream would be typing just 'trampolino --corpus-callosum' and getting the tractogram for a corpus callosum to play with!
More information in this github issue

Neuroimaging tutorials and resources: A list of tutorials and other resources useful to learn open science and neuroimaging
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
The purpose of this project is to gather courses, software and any resources that can help any newcomer (e.g. neuroscientist, computer scientist). In other words, if you wonder Where can I find a good: Tutorial on X? Software to do Y?, we hope that this project will answer your question. See this link for more info.
More information in this github issue

Python library to handle confounds/covariate
Hub : Americas
PROJECT DESCRIPTION
Develop a python library of methods to handle confounds in various neuroscientific analyses, esp. statistics and predictive modeling.
More information in this github issue

Building 3R-BRAIN Neuroinformatics
Hub : Asia / Pacific
PROJECT DESCRIPTION
An Open Resource for Reproducible, Replicable and Reliable Brain Research and Imaging Neuroscience.
During the hack time, we are going to build a neuroinformatic resource for the 3R-BRAIN data sharing. This includes the resources for OHBM education course 'Population Neuroimaging: How to responsibly handle big data in the age of biobanks', sharing big data of 3R-BRAIN, and the implementation of reliability analysis toolkit into an online connectome computation system (CCS).
More information in this github issue
Prediction of age and intelligence, a basic machine learning model
Hub : Asia / Pacific
PROJECT DESCRIPTION
Emerging evidence suggests the outstanding utility of multimodal neuroimaging data in predicting individual variation in age and cognition. However, it is unclear whether the predictive utility of such measures is a global effect or is consistently attributed to certain localized patterns of brain structure and connectivity.
This project aims to evaluate and develop an analysis framework for optimal performance of age and behavior (e.g. fluid intelligence) prediction using measures of brain structure and connectivity. The goal is to achieve both overall high prediction accuracy in cross-validated samples and high consistency in the estimated predictors (e.g. regions/connections) if possible. A secondary aim of the project is to investigate the potential markers of sub-clinical cognitive decline through simultaneous prediction of age and cognition in a healthy adult cohort.
More information in this github issue
Virtual Neuro Machine (VNM)
Hub : Asia / Pacific
PROJECT DESCRIPTION
With the advent of container technology, cumbersome virtual machines have been reduced to a single container file that can be easily run on multiple-environments. Containers can be run both interactively (e.g., within a Virtual Machine on the cloud) and in batch processing (e.g., on HPC), thus contributing to reproducibility across stages of the analysis. The goal of this project is to generate a 'virtual neuro machine' container, i.e. a large container that includes most popular neuroimaging tools, and can be used as a stand-alone VM. Although tools to generate neuroimaging software containers are already in place (e.g., neurodocker), continued maintenance of such a large container with complex library dependencies requires in-built testing capabilities, and especially so, of included GUI applications. This project will bring together people working on different environments and using different neuroimaging tools, in order to 1) build a working container with the newest software versions available in June 2020, 2) develop in-built testing capabilities, to facilitate future container upgrades, 3) develop a standard 'home screen' for the container, to provide common interface across host environments.
More information in this github issue

BIDS-prov examples
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
Provenance has already been discussed in the bids specification, see this commit. Adding more examples in the form of json-ld files, would allow users to better picture workflows made possible by this.
More information in this github issue

BIDS Specification Terms PDF Table Generator
Hub : Americas
PROJECT DESCRIPTION
The project aims to create a user-friendly python tool allowing the BIDS group to select, search, or add BIDS specification terms and export a PDF table of the chosen BIDS term along with appropriate term properties and instructions on adding new terms. If the user chooses to add a new term, the tool will create and export a new BIDS specification term as a JSON-LD file and push it to our terms repository where discussion surrounding the new term can be captured using git/github. Such an effort will provide the BIDS working group with a user-friendly tool to build tables for the BIDS documentation while ensuring they have not re-purposed an existing BIDS specification term giving it conflicting definitions. For additional information on the background for this project see BIDS specification issue 423.
More information in this github issue
HyPyP – The Hyperscanning Python Pipeline
Hub : Europe / Middle-East / Africa
PROJECT DESCRIPTION
HyPyP is the first open source toolkit for the analysis of hyperscanning recording i.e. neural activity across multiple brains. Here we provide a Python library based on the MNE Python ecosystem.
More information in this github issue

Sharing datasets and pipelines in the Canadian Open Neuroscience Platform
Hub : Americas
PROJECT DESCRIPTION
The Canadian Open Neuroscience Platform (CONP) is a web portal and DataLad repository to share open and restricted-access datasets and tools in the neuroimaging community. Datasets are uniformly described using the Data Tags Suite (DATS), available for download through DataLad, regularly monitored using CircleCI, and searchable through the CONP web portal. The data itself remains under the control of its owners, as it can be stored in various backends including LORIS, Zenodo, the Open Science Framework, or any other backend supported by DataLad (http, ftp, etc). Tools are shared on Zenodo through the Boutiques framework. The goal of this project is to port additional tools and pipelines to the CONP. Please join us if you would like to share your dataset, your pipeline, or just learn about the technologies!
More information in this github issue
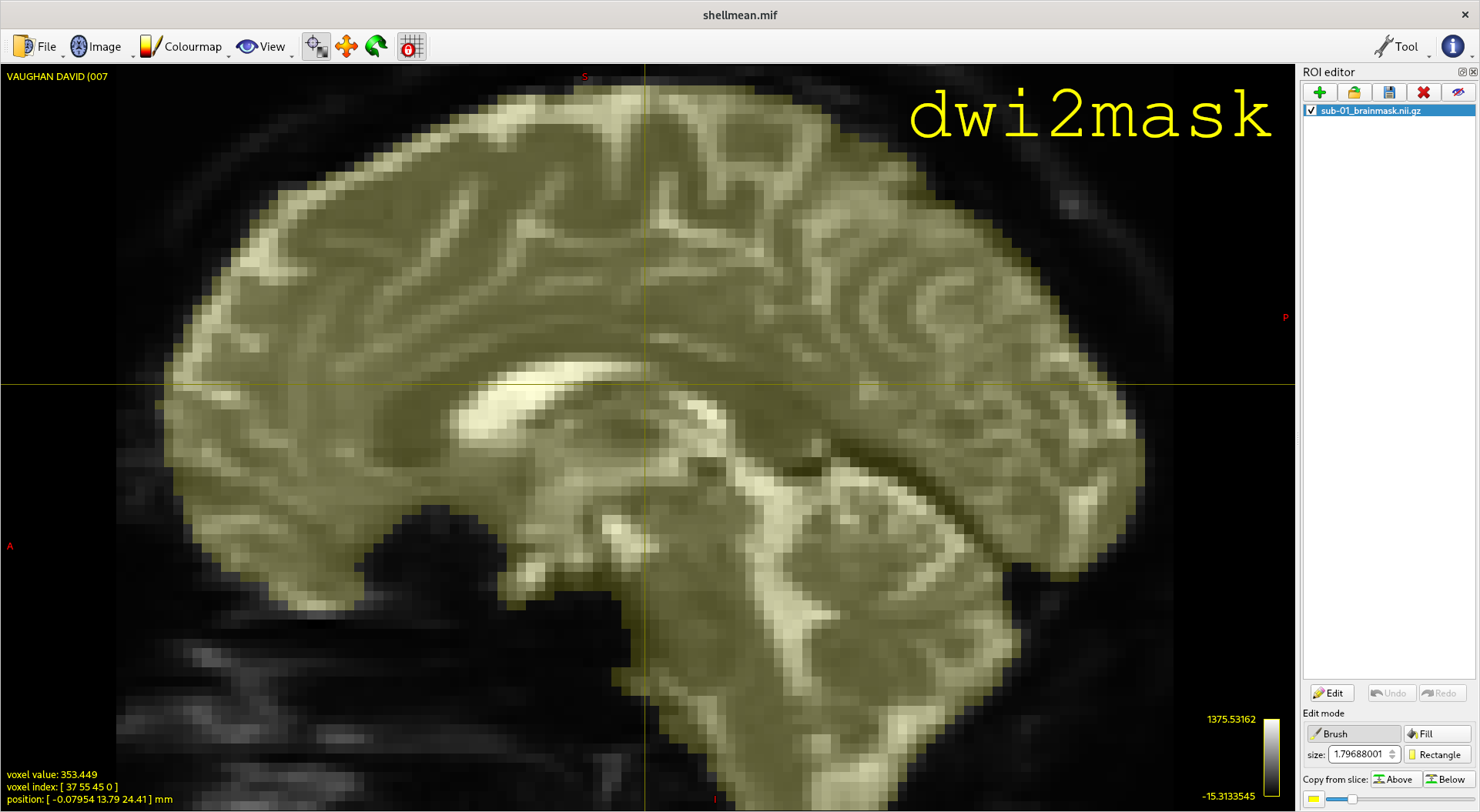
Diffusion MRI brain masking ("dwi2mask")
Hub : Asia / Pacific
PROJECT DESCRIPTION
Deriving a binary brain mask from diffusion MRI data in an automated fashion can be deceptively challenging. This project aims to provide a standardised interface to multiple different algorithms for performing this task, and incorporate it into the MRtrix3 software.
More information in this github issue
BIDS Curation GUI
Hub : Americas
PROJECT DESCRIPTION
Getting data into the Brain Imaging Data Structure (BIDS) format is non-trivial. Ideally, you set scan acquisition parameters in preparation for BIDS, but mistakes will be made. Non-ideally, you have an old set of scans you want to BIDSify. Wouldn't it be great if you could 'see' the entire situation so you can easily locate and understand the problems? Wouldn't you like this picture to come alive to help you curate your data?
More information in this github issue

ReproSchema and Library: A JSON-LD schema to harmonize behavioral, cognitive, and neuropsych assessments
Hub : Americas
PROJECT DESCRIPTION
Harmonizing data after acquisition is resource intensive. ReproSchema is an attempt to harmonize data by design by enforcing consistency at the data acquisition stage. An implementation of the schema can specify scoring logic, branching logic, and user interface rendering options. The schema allows internationalization (multiple languages), is implementation agnostic, and tracks variations in assessments (e.g., PHQ-9, PHQ-8).
This open and accessible schema library with appropriate conversion (e.g., to RedCap) and data collection tools (e.g., MindLogger, LORIS, RedCap) enables more consistent acquisition across projects, with results being harmonized by design.
More information in this github issue
Pyrda's relatives and friends: providing Pydra's support for FitLins, Nipype interfaces and ML model comparison
Hub : Americas
PROJECT DESCRIPTION
Pyrdais a lightweight, Py3.7+ dataflow engine for computational graph construction, manipulation, and distributed execution. Pydra is a part of the second generation of the Nipype ecosystem, but it is designed as a general-purpose engine to support analytics in any scientific domain.
FitLins is a tool for estimating linear models, defined by the BIDS Stats Models specification proposal, to BIDS-formatted datasets. is a demo application that leverages Pydra together with scikit-learn to perform model comparison across a set of classifiers.
More information in this github issue
physiopy/phys2bids: BIDS formatting of physiological recordings
Hub : Americas & Europe / Middle-East / Africa
PROJECT DESCRIPTION
Physiopy is a github organisation that aims to offer open development solutions for physiology in MRI. phys2bids is the flagship library of physiopy, and it consists in a python3 library meant to format physiological (cardiac, respiratory, or gas signals) files in BIDS.
During this hackathon, we would love to involve new contributors to integrate new features, such as automatic recognition of the file content, support for eyetracking, and integration with other bidsificators (such as bidscoin) Next to phys2bids, we’re starting the development of phys2denoise, a library dedicated to prepare physiological signal for fMRI denoising.
More information in this github issue

Creating a dashboard and recommender system for CONP DataLad datasets and Boutiques pipelines
Hub : Americas
PROJECT DESCRIPTION
Our ultimate goal is to implement a dashboard and recommender system for Boutiques pipelines and DataLad datasets, in the context of the Canadian Open Neuroscience Platform (CONP). In this project we will focus on implementing a database to track information on pipelines, datasets and available execution records. Using this database, we will extract useful information for this recommendation system.
More information in this github issue

PyNets
Hub : Americas
PROJECT DESCRIPTION
PyNets leverages the Nipype workflow engine, along with Nilearn and Dipy fMRI and dMRI libraries, to sample individual structural and functional connectomes. Uniquely, PyNets enables the user to specify any of a variety of methodological choices (i.e. that impact node and/or edge definitions) and sampling the resulting connectome estimates in a massively scalable and parallel framework. PyNets is a post-processing workflow, which means that it can be run manually on virtually any preprocessed fMRI or dMRI data. Further, it can be deployed as a BIDS application that takes BIDS derivatives and makes BIDS derivatives.
More information in this github issue
Matching white matter fiber tracts across subjects
Hub : Asia / Pacific
PROJECT DESCRIPTION
With diffusion MRI, we can track the axon fibers in the brain white matter. Although a certain level of variability exists, we still believe that there are a substantial amount of fiber bundles/tracts that are common across subjects. Thus, matching the fiber bundles across subjects could help us understand the common architecture of white matter in the human brain.
More information in this github issue
BIDS Connectivity data schema
Hub : Europe / Middle East / Africa
PROJECT DESCRIPTION
The Generic BIDS connectivity data schema is a BIDS proposal submitted to explore the development of standards for representing connectivity data BIDS derivative schemas across modalities. This would enable easier integration of multimodal data in tools, and ease the expansion of BIDS to cover more modalities. Now derivative schemas across more modalities are becoming more mature, and simpler elements of data representation have been addressed, now is a good time to review the status of connectivity data representation and determine what a 'meta' schema might be able to provide in this context.
More information in this github issue
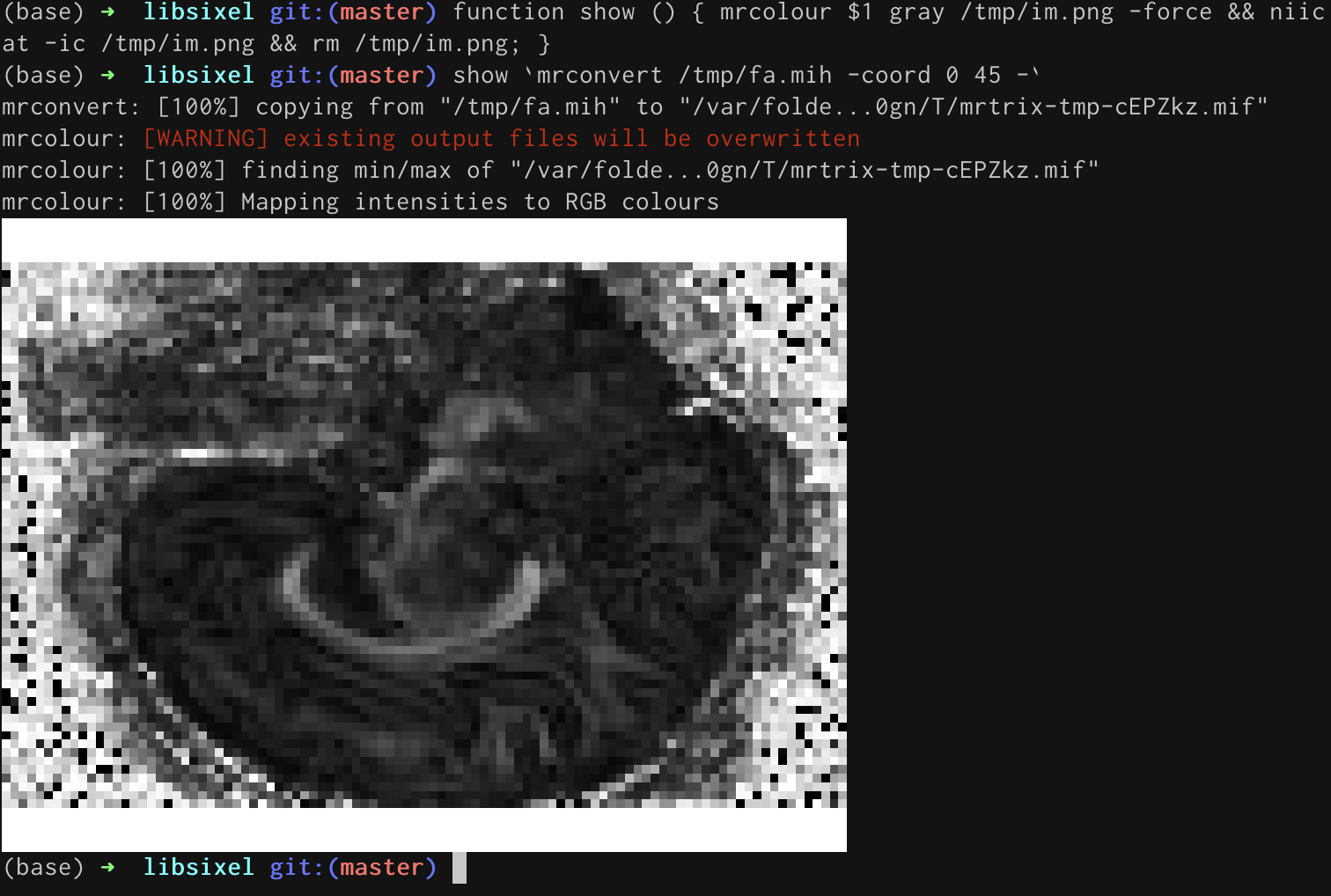
mrpeek: a sixel-based terminal image viewer for MRtrix3
Hub : Europe / Middle East / Africa
PROJECT DESCRIPTION
Displaying images on a remote server is very difficult / impossible currently using MRtrix3's mrview, due the incompatibility of OpenGL 3+ with X11 forwarding (see here for details). It will be some time before the technology is updated to fix this. In the meantime, @wasserth's niicat project shows that it is possible to allow some (limited) display functionality using sixel. The libsixel project provides an implementation that seems like a good starting point for a C++ command.
The aim of this project is to create a simple command for interactive display of MR images within the terminal, build on the MRtrix3 codebase, so that it can be used for quick inspection of images both locally and remotely. If successful, changes will be included in a future update to the MRtrix3 software. Contributors will be included in the built-in git contribution statistics. We additionally credit contributors by showing their avatar at the bottom of the MRtrix3 website front page, and in the changelog as reported on the community forum (example).
More information in this github issue
Evaluating the pipeline stability though the libmath fuzzy environment
Hub : Americas
PROJECT DESCRIPTION
Data analysis pipelines are known to be impacted by the operating system updates, presumably due to the creation, propagation, and amplification of numerical noises. These are uncontrolled noises that are originated in updates of low-level libraries by third-party developers. In practice, these numerical instabilities are caused by truncation and round-off errors due to the precision limitation of the floating-point arithmetic. The finite precision of the floating-point numbers leads to represent an approximation of a real number that may be different on the computers with various precision formats. A possible method to investigate operating system related effects more comprehensively is to introduce controlled numerical perturbations in the math library functions (libmath), which could be done by introducing noise in floating-point computations through Monte-Carlo Arithmetic (MCA) using the Verificarlo tool.
For the hackathon we'll be looking to test the validity of the instrumented libmath library functions and also evaluate the stability of the existing neuroimaging pipelines.
More information in this github issue

Building Citizen Science Quality Control Exploratory Data Analysis & Other Tools
Hub : Americas
PROJECT DESCRIPTION
Swipes For Science is a citizen science game framework brought to life by Anisha Keshavan, PhD, et al, inspired by Tinder. Citizen scientists swipe left or right to judge an image as Pass or Fail and each vote is recorded into a live online database. We have a unique opportunity at this OHBM Brainhack 2020 for you to both setup your own Swipes For Science site and to create exploratory data analysis tools with anonymized data collected from an existent Swipes For Science site. You can imagine answering questions like:
- For all the images associated with one subject's data, should that entire subject be considered a Pass or Fail for inclusion in further analysis?
- For all ratings provided by citizen scientist ABC, how reliably does ABC vote compared to the gold standard data?
- Among all citizen scientists, who are the best raters?
- What is the inter-rater reliability on image XYZ?
... and more!
More information in this github issue
Integration of EEG in the Connectome-Mapper 3
Hub : Europe / Middle East / Africa
PROJECT DESCRIPTION
Flexible pipelines for both diffusion MRI and functional MRI have been implemented in different software such as the Connectome Mapper 3, an open-source pipeline software, released as a BIDS App, for mapping hierarchical multi-scale connectomes from multi-modal datasets, but solutions for EEG and MEG are still lacking. This project intents to extends Connectome Mapper 3 to EEG.
More information in this github issue
About the Open-Science Special Interest Group
The OHBM Open Science SIG mission is to advance neuroimaging research by fostering the open sharing of ideas, data, and tools between members of the OHBM community. This will be accomplished by organizing educational events, supporting collaborative initiatives, and providing mentorship to junior researchers.As with previous years, our three main contributions to the OHBM annual meetings are:
- organizing a Brainhack to precede the annual meeting
- introducing the tools of open science via interactive tutorials as part of the TrainTrack
- facilitating collaborative work on a wide range of open science projects
This year the event is happening virtually, and we look forward to this opportunity to reach out to a larger and more diverse audience and to introduce a number of innovations that will enhance virtual collaborations!